Graph Syntax
FGUtils has its own graph description language. The syntax is closely related
to the SMILES format for molecules and reactions. It is kind of an extenstion
to SMILES to support modeling ITS graphs and reaction patterns. To convert the
SMILES-like description into a graph object use the
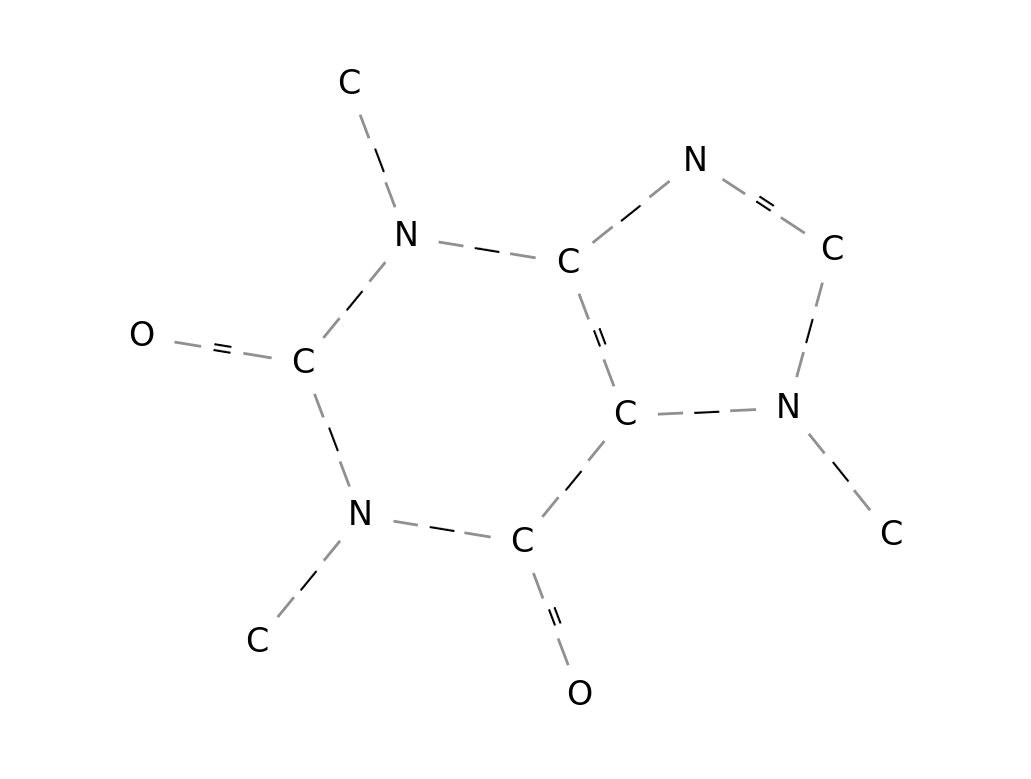
Parser class. The Caffeine molecular graph can be
obtained as follows:
import matplotlib.pyplot as plt
from fgutils import Parser
from fgutils.vis import plot_as_mol
parser = Parser()
mol = parser("CN1C=NC2=C1C(=O)N(C(=O)N2C)C")
fig, ax = plt.subplots(1, 1)
plot_as_mol(mol, ax)
plt.show()
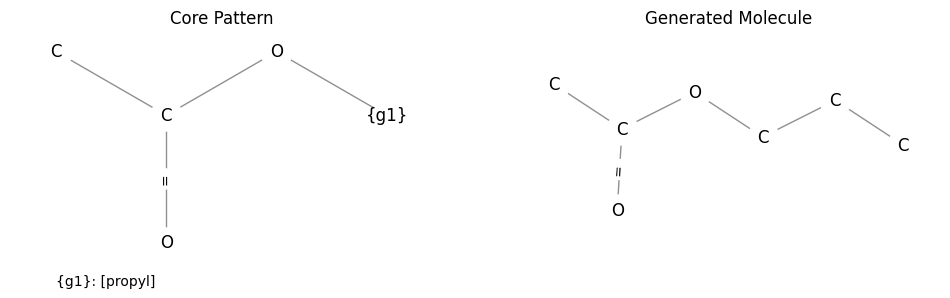
Besides parsing common SMILES it is possible to generate molecule-like graphs
with more abstract nodes, i.e., arbitrary node labels. Arbitrary node labels
are surrounded by {} (e.g. {label}). This abstract labeling can be used
to substitute nodes with specific patterns. In this context the labels are
group names of ProxyGroup objects. A ProxyGroup
defines a set of sub-graphs to be replaced for the labeled node. This can be
done by using a Proxy. Propyl acetate can be created
by replacing the labeled node with the propyl group:
import matplotlib.pyplot as plt
from fgutils import Parser
from fgutils.proxy import MolProxy, ProxyGroup
from fgutils.vis import GraphVisualizer
pattern = "CC(=O)O{propyl}"
propyl_group = ProxyGroup("propyl", "CCC")
parser = Parser()
proxy = MolProxy(pattern, propyl_group, parser=parser)
g = parser(pattern)
mol = next(proxy)
vis = GraphVisualizer()
fig, ax = plt.subplots(1, 2, dpi=100, figsize=(12, 3))
vis.plot(g, ax[0], title="Core Pattern")
vis.plot(mol, ax[1], title="Generated Molecule")
plt.show()
Note
A node can have more than one label. This can be done by separating the
labels with a comma, e.g.: {label_1,label_2}.
In the example above the ProxyGroup has only one subgraph pattern. In general,
a ProxyGroup is a collection of several possible subgraphs from which one is
selected when a new sample is instantiated. To get more information on how
graphs are sample take a look at the GraphSampler
class and the ProxyGroup constructor. By default a
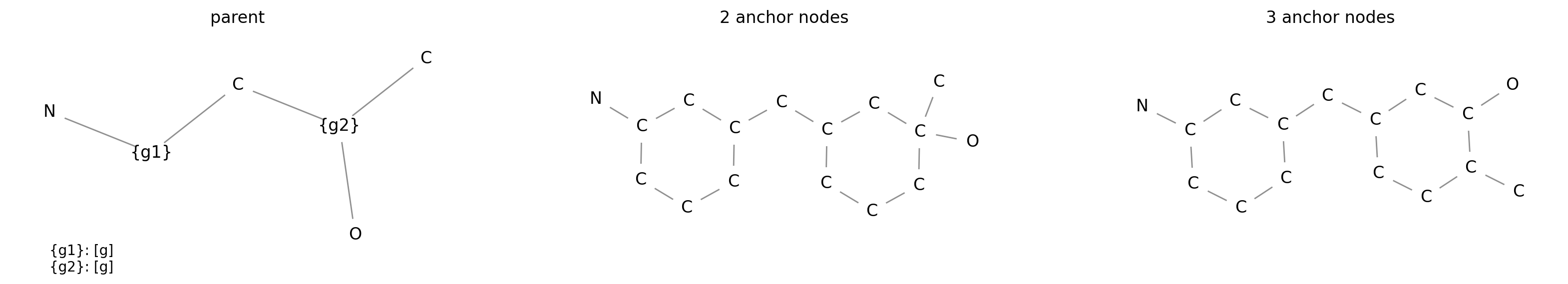
pattern has one anchor at index 0. If you need more control over how a subgraph
is inserted into a parent graph you can instantiate the
ProxyGraph class. For a ProxyGraph you can provide a
list of anchor node indices. The insertion of the subgraph into the parent
depends on the number of anchor nodes in the subgraph and the number of edges
to the labeled node in the parent. The first edge in the parent connects to the
first anchor node in the subgraph and so forth. The following example
demonstrates the insertion with multiple anchor nodes:
import matplotlib.pyplot as plt
from fgutils.proxy import MolProxy, ProxyGroup, ProxyGraph, Parser
from fgutils.vis import GraphVisualizer
pattern = "N{g}C{g}(O)C"
g_1 = ProxyGroup("g", ProxyGraph("C1CCCCC1", anchor=[1, 3]))
g_2 = ProxyGroup("g", ProxyGraph("C1CCCCC1", anchor=[1, 3, 4]))
parser = Parser()
proxy1 = MolProxy(pattern, g_1)
proxy2 = MolProxy(pattern, g_2)
parent_graph = parser(pattern)
mol1 = next(proxy1)
mol2 = next(proxy2)
vis = GraphVisualizer(show_edge_labels=False)
fig, ax = plt.subplots(1, 3, dpi=200, figsize=(20, 3))
vis.plot(parent_graph, ax[0], title="parent")
vis.plot(mol1, ax[1], title="2 anchor nodes")
vis.plot(mol2, ax[2], title="3 anchor nodes")
plt.show()
Another extension to the SMILES notation is the encoding of bond changes. This
feature is required to model reaction mechanisms as ITS graph. Changing bonds
are surrounded by <> (e.g. <1, 2> for the formation of a double bond
from a single bond). The extended notation allows the automated generation of
reaction examples with complete atom-to-atom maps. The following code snippet
demonstrates the generation of a few Diels-Alder reactions. The diene and
dienophile groups can of course be extended to increase varaity of the
samples:
import random
import matplotlib.pyplot as plt
from fgutils.proxy import ProxyGroup, ProxyGraph, ReactionProxy
from fgutils.proxy_collection.common import common_groups
from fgutils.vis import plot_reaction, plot_its
from fgutils.its import get_its
electron_donating_group = ProxyGroup(
"electron_donating_group",
["{methyl}", "{ethyl}", "{propyl}", "{aryl}", "{amine}"],
)
electron_withdrawing_group = ProxyGroup(
"electron_withdrawing_group",
["{alkohol}", "{ether}", "{aldehyde}", "{ester}", "{nitrile}"],
)
diene_group = ProxyGroup(
"diene",
ProxyGraph("C<2,1>C<1,2>C<2,1>C{electron_donating_group}", anchor=[0, 3]),
)
dienophile_group = ProxyGroup(
"dienophile",
ProxyGraph("C<2,1>C{electron_withdrawing_group}", anchor=[0, 1]),
)
groups = common_groups + [
electron_donating_group,
electron_withdrawing_group,
diene_group,
dienophile_group,
]
proxy = ReactionProxy("{diene}1<0,1>{dienophile}<0,1>1", groups)
n = 4
fig, ax = plt.subplots(n, 2, width_ratios=[2, 1], figsize=(20, n * 4))
for i, (g, h) in enumerate(random.sample(list(proxy), n)):
plot_reaction(g, h, ax[i, 0], title="Reaction")
plot_its(get_its(g, h), ax[i, 1], title="ITS Graph")
plt.tight_layout()
plt.show()
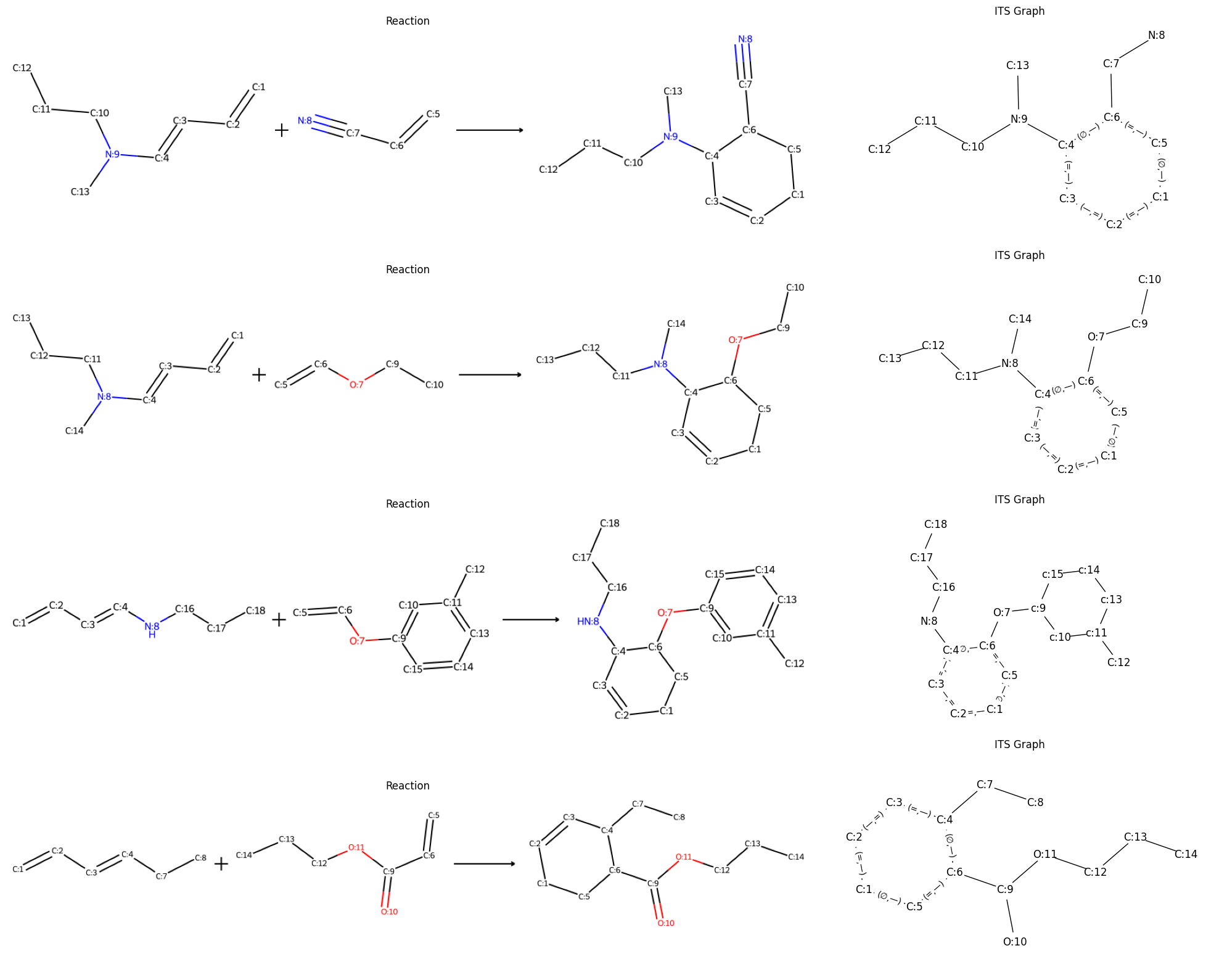
This proxy can now generate Diels-Alder reaction samples. A few of the results
are shown in the figure above. On the left side the reaction and on the right
side the resulting ITS. The results are balanced and have complete atom-to-atom
maps. The atom-to-atom maps are correct as long as the configuration makes
sence in the chemical domain. Note that the synthesizability of the generated
samples can not be guaranteed. It soley depends on what ProxyGroups and
ProxyGraphs are configured. For a comprehensive Diels-Alder reaction proxy take
a look at the
DielsAlderProxy class
and the section TODO. This class is also able to generate negative Diels-Alder
reaction samples, i.e., reactions where a Diels-Alder graph transformation rule
is theoretically applicable but the reaction will never happen in reality.
Note
The electron_donating_group and electron_withdrawing_group serve as
a collection of other groups to simplify the notation. They consist of a
single node with multiple labels. When iterating the next sample from the
proxy the labeled nodes get replaced by the pattern from one of the groups.
The group/label is chosen randomly with uniform distribution.
Warning
The call list(proxy) will generate all possible instantiations at once.
Depending on the configuration this can take a long time to complete. If the
core ProxyGroup graph sampling is not unique this can even result in an
endless loop.